Machine Learning Engineer
Machine Learning Engineer
Maciej Wiatrak explains the work he presented at the LOUHI Workshop on Health Text Mining and Information Analysis at EMNLP 2022 and its implications for entity linking in biomedical text analyses.
The meaning of a single term that one may encounter in a text depends heavily on its context. For example, the word expression may mean facial expression, gene expression or even just the act of articulating feelings. Humans are good at discerning the meaning of words given the context in which they are used, but this task is a challenge for machines. Considerable time has been spent on AI/ML algorithms for grounding biomedical terms to their actual meaning, also known as the entity linking task. Entity linking is one of the crucial first steps for a number of biomedical text analysis tasks performed at BenevolentAI, such as relation extraction and target identification.
Entity linking algorithms leverage context to assign a textual mention to its corresponding entity. In the example above the algorithm may reason that, if the text surrounding word expression talks about genes, the mention should be assigned to the gene expression rather than the facial expression entity. Each textual mention that was independently recognised as interesting needs to be assigned to a corresponding entry in the knowledge base, where the knowledge base is usually a manually curated collection of entities. Knowledge bases are often very large; for example the Unified Medical Language System (UMLS) [1], curated by the National Institute of Health, contains millions of unique entities such as diseases, genes, pathways and many others. Comparing each mention to all of the entities in such a knowledge base could take a lot of time and computational power. Additionally, knowledge bases can rapidly change over time, particularly in the biomedical domain.
In this work presented at the EMNLP LOUHI workshop, we developed an entity linking system that can accurately link a term to its entry in the knowledge base without comparing it to every entity during the computationally expensive training phase. This makes the process much simpler and faster. We accomplished this by designing a new proxy-based entity linking loss function, which is more performant and scalable compared to the standard cross-entropy loss function when dealing with large knowledge bases.
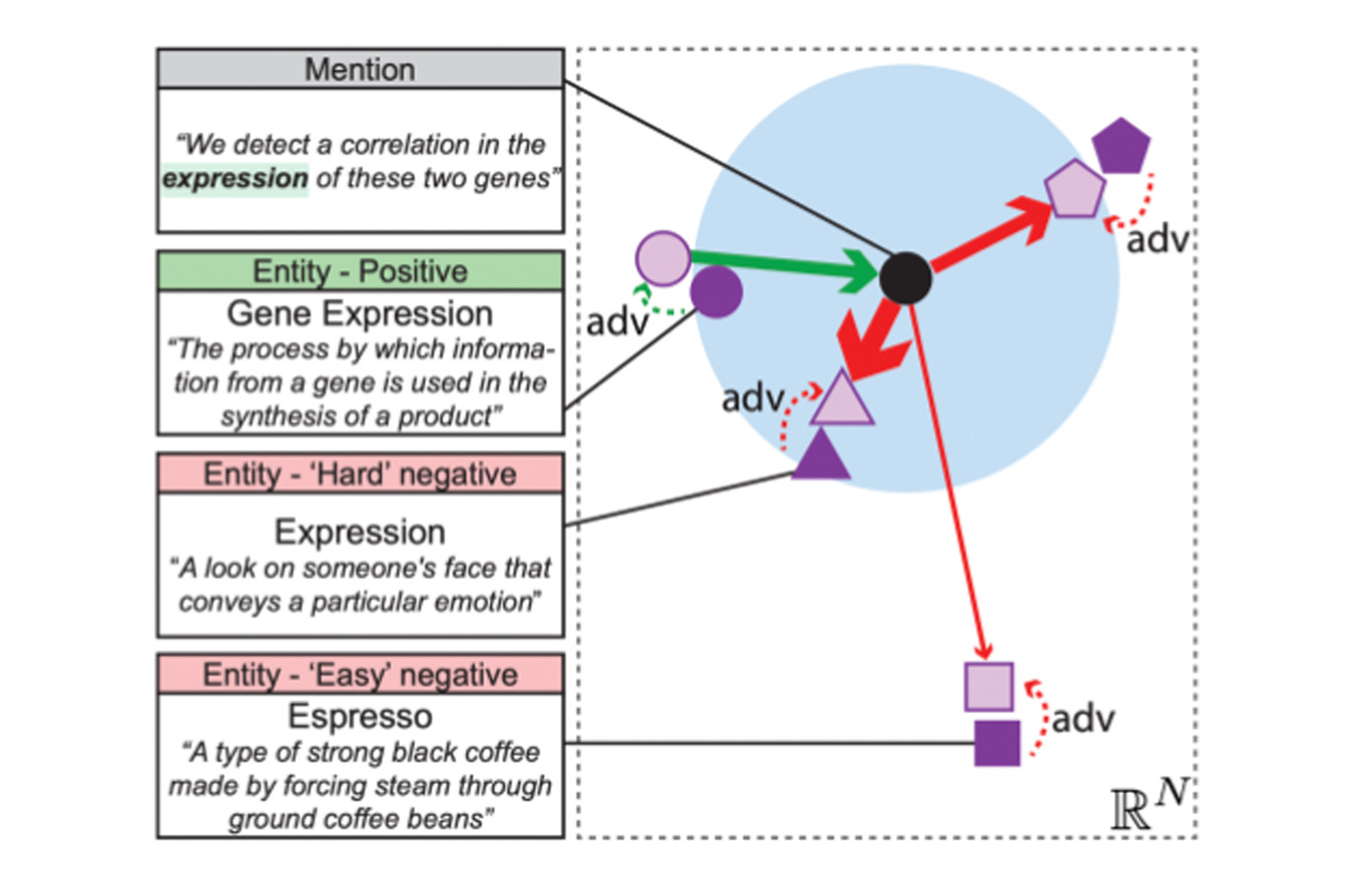
Figure 1: Overview of our proxy-based entity linking method. The entities are “pushed” and “pulled” away depending on how difficult it is for the model to discern what is the right entity for a mention.
Another issue that we tackle in our work is the discovery of out-of-knowledge-base entities. Sometimes, an entity linking system encounters a mention that does not fit any of the entries in the knowledge base. In such a situation, the system has two choices: 1) flag it for manual evaluation if it is potentially interesting or 2) discard it as an irrelevant mention. In our paper, we show that our proxy-based entity linking loss can efficiently discern between the two, while simultaneously retaining high overall accuracy. This allows us to easily expand the knowledge base by discovering and flagging interesting mentions that are not yet in the knowledge base.
Together, these features allow us to scalably and efficiently perform entity linking, and also to potentially find new, relevant entities that we can add to the knowledge base. At BenevolentAI we regularly process millions of scientific articles, and making sense of them is a key component of our approach. Getting entity linking right is crucial for the downstream text analysis tasks. We hope that applications of this novel method in biology bring us one step closer to discovering life-changing medicines with a higher likelihood of clinical success.
References
[1] Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D267-70. doi: 10.1093/nar/gkh061. PubMed PMID: 14681409; PubMed Central PMCID: PMC308795.
Back to blog post and videos