Adam Foster recently presented a paper at ICML 2022 based on work that he did during his AI Internship at BenevolentAI. Below, Sam Abujudeh and Aaron Sim, who also contributed to this research, explain more about the work and its implications for data integration, counterfactual inference and fairness in machine learning.
Machine learning and the challenges of biological data
Alongside the steady stream of breakthroughs in artificial intelligence, there is a quieter revolution taking place in molecular biology. This is, of course, the increasingly remarkable ability to look inside a cell through the lens of omics data. It is tempting to believe that these two fields are made for each other. After all, the insatiable data requirements of modern machine learning algorithms are well served by the ever-growing store of omics datasets, from single-cell transcriptomics to genome-scale CRISPR interventional data with millions of data points. In return, the machine learning subfield of representation learning is practically tailor-made for computational biologists whose daily workout consists of turning high-dimensional data (e.g. gene expression) into low numbers of interpretable and biologically meaningful features for all manner of useful applications from drug target selection to patient clustering in precision medicine.
However, dig a bit deeper and the familiar challenges of biological data remain. For a start, omics is not so much Big Data as it is a frustratingly heterogeneous collection of small studies that resists all simple attempts at integration. And even in larger datasets without ‘batch effects’, the biological context is often different. For instance, we make do with accessible cell lines rather than actual tumour samples, or accept that a biobank’s participant demographics are not representative of the population at large. In our paper, we propose a method we call Contrastive Mixture of Posteriors, or CoMP for short, that addresses these data challenges en route to learning representations of data with a few surprisingly powerful downstream applications.
Marginal independence of representations and conditions
We start by separating the raw data collected by an experimental apparatus from its accompanying conditions, such as the information on the experimental methods employed, the source of the data, or the gender of the study participants, etc. Within a probabilistic framework, we identify that the common symptom of the data problems highlighted above is the fact that while we would like to safely ignore these condition variables, for example analysing cell line data as though it comes directly from patient samples, the representations we learn and work with are not independent of them. Our approach is to directly enforce the marginal independence of these two variables.
To do this we take inspiration from contrastive representation learning. In contrastive learning, one learns to pull representations of ‘similar’ data points close to one another while pushing ‘dissimilar’ points far apart. The key idea in CoMP is this: we simply nudge our model, via a simple loss function penalty, to treat all pairs of data points with, or generated under, different conditions as similar, and pairs with identical conditions as dissimilar. Working in a conditional variational autoencoder framework, we can make concrete the notion of close vs. far directly in terms of the probability distributions that the model itself defines. As we can see in the figure below our method achieves a visually pleasing overlap of conditions while retaining the identifiability of relevant biological features.
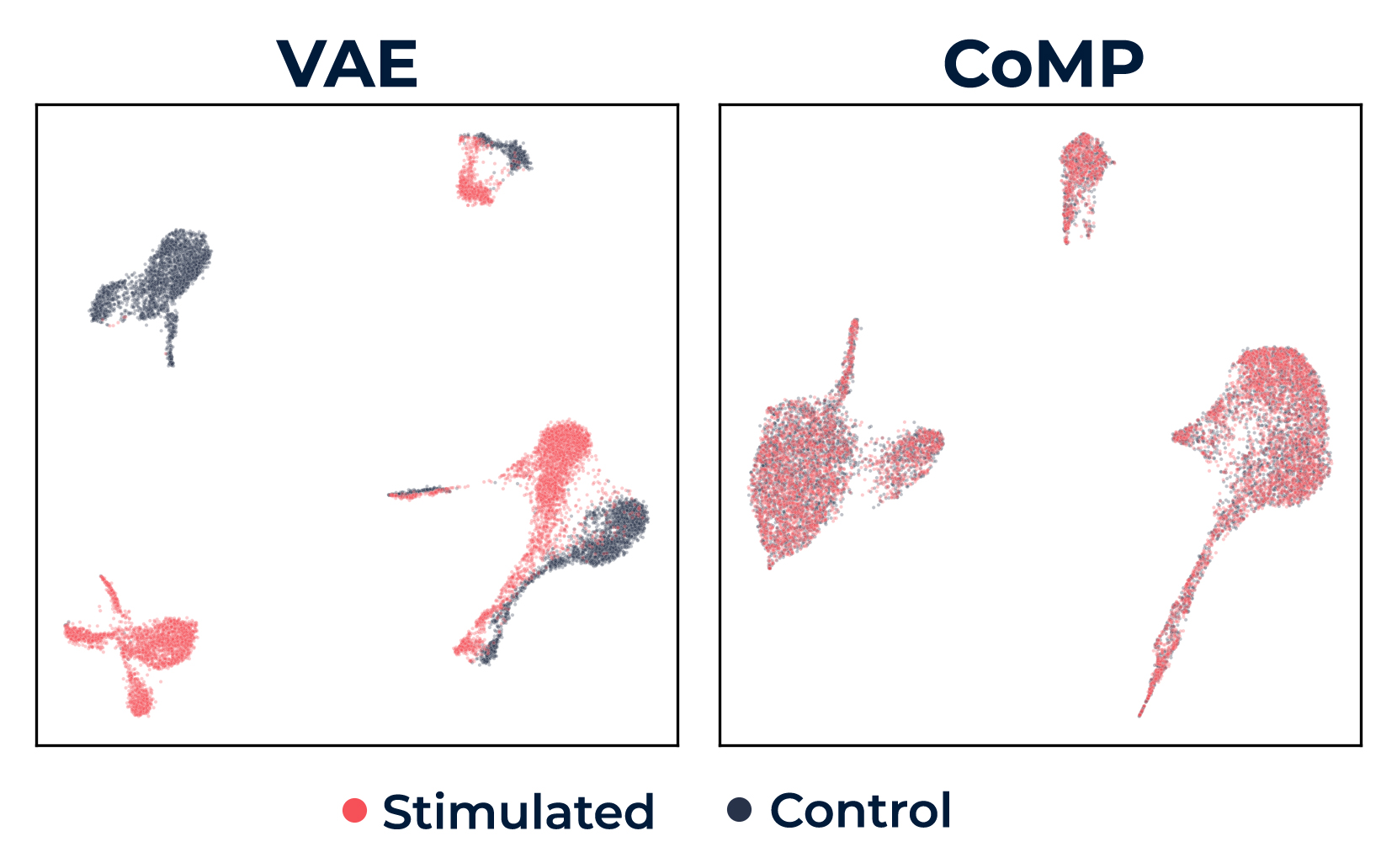
Figure 1. Latent representations of a single-cell gene expression dataset under two conditions: stimulated (red) and not stimulated (black), showing fully disjointed (VAE) on the left to a well-mixed pair of distributions (CoMP) on the right.
Fair representations and counterfactual inference
While the removal of batch effects in omics data and improving data integration is an obvious benefit of CoMP, learning data representations that are marginally independent of a condition variable has two important applications that extends outside of these applications and even outside of biology. First, CoMP allows one to effectively censor a given attribute from our data to learn ‘fair’ representations. For instance, AI systems operating on these representations can now be employed safely in the knowledge that protected attributes – such as ethnicity, gender or age – are not factors in any automated decision making; achieving this without significant loss in representation power is an important step towards the broader goal of ethical AI. Second, because CoMP produces representations independent of the condition, we can now perform counterfactual inference by flipping our condition variable and decoding the result in silico, to ask, for example, how would the expression of genes in a given cell change if treated with a certain drug. Such “what if” questions are a large part of what early drug discovery is all about.
PUBLICATION
Contrastive Mixture of Posteriors for Counterfactual Inference, Data Integration and Fairness
Back to blog post and videos