BenevolentAI has released GuacaMol, a framework to benchmark models for de novo molecular design. Researchers are encouraged to participate in the competition.
The needle in the haystack
Many novel materials or new drugs involve molecules that cannot be found in nature. Designing such molecules is a complicated but thrilling task: since the number of possible molecules can be considered to be, for practical purposes, infinitely large, it is a challenge to identify novel molecules with the required properties before they have ever been synthesised. Traditionally, experienced chemists apply their knowledge to iteratively improve molecules. For instance, they are able to identify structural motifs or functional groups and estimate their effect on the properties of a molecule.For a few decades, researchers both in industry and academia have been designing computational models to assist chemists in tackling this complex task. These models propose new molecules to synthesise, with the expectation that they will exhibit the desired properties. This computational technique is commonly called “de novo design” (and one often refers to the corresponding models as “generative models”).In the last couple of years, many new generative models relying on artificial intelligence (AI) have been proposed - the absence of exact rules in molecular design, or the difficulty to formulate them, usually indicates that AI models should do better than rule-based models. These new models have already proven to be beneficial in several applications. At BenevolentAI, we employ generative models to enhance drug discovery - we synthesise what our computational models suggest to augment what our medicinal chemists design.
Complicated models are not necessarily better than simpler ones, so what to choose?
The downside to the increasing interest in de novo design is that it becomes difficult to maintain an overview of what models exist at all, and to know which ones perform better than the others. As Ockham’s razor tells us, complicated models are not necessarily better than simpler ones: possibly, complex AI models may not be the best ones.At the same time, it is hard to define tasks to compare models, because there is not one single way to tell whether a model is good or bad. This explains why there has been no consistency in the metrics evaluated by the proposed models until now. To drive progress, it is essential to allow for a direct and fair comparison between generative models - or, in other words, to define standardised benchmark for de novo design. In other fields, such as image recognition, standardised tasks formulated as competitions have led to rapid progress. Is the same thing possible for de novo molecular design?
Our answer: Guacamol
In a recent publication, we tackled this challenge. We defined a series of tasks that generative models should be able to fulfil, at least to some extent, and that allow us to measure their strengths and weaknesses with respect to different criteria. For this endeavour, an important decision was to distinguish between two categories of tasks commonly tackled by generative models: first, the ability to mimic molecules from a reference set, and, second, the ability to generate molecules with specific property profiles. To reflect this, we defined 5 distribution-learning benchmarks and 20 goal-directed benchmarks, respectively.
To simplify the assessment of new generative models, we implemented GuacaMol, an open-source Python package, that was designed with the intention to allow researchers to test their new models as easily as possible.With the help of GuacaMol, we evaluated a series of baselines generative models selected from many recently proposed methods. Assessing baselines models provides interesting insight into their strengths and weaknesses, and highlights the differences and similarities between them. Also, GuacaMol automatically saves the molecules generated during the evaluation, as their inspection may illustrate the peculiarities of the model capabilities.Scores of the baseline models on the distribution-learning tasks.
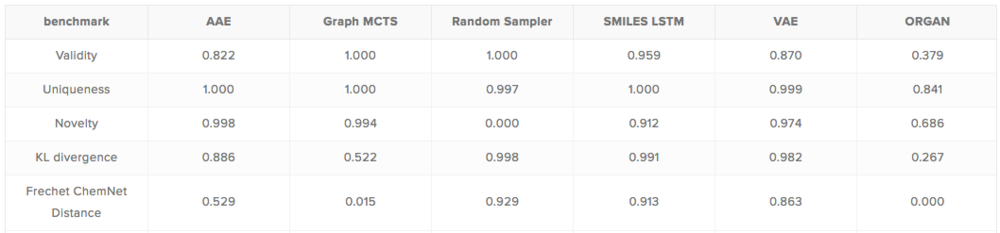
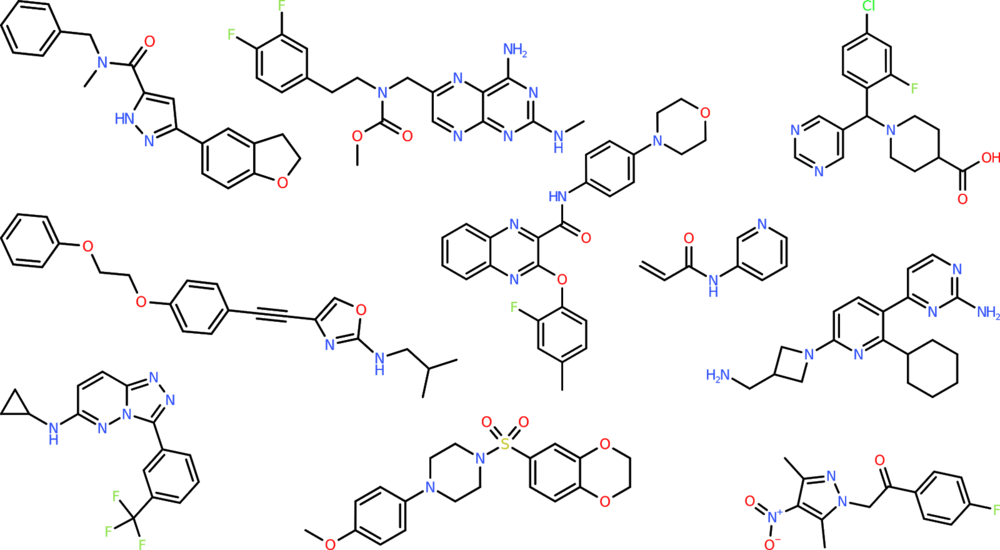
As the results for distribution-learning tasks show, SMILES LSTM, an AI model with roots in language modelling, is currently the most suitable model for mimicking reference sets. The figure below shows shows 10 molecules generated by this model (more can be found on http://benevolent.ai/guacamol).
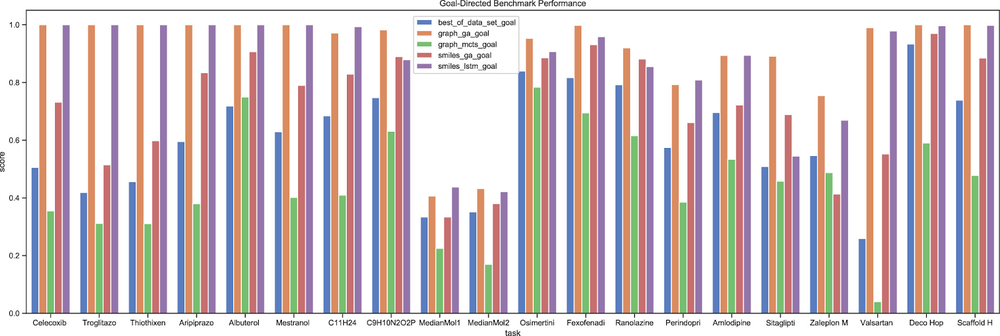
For the goal-directed benchmarks, genetic algorithms (GAs) and neural models run head-to-head, with the graph-based GA model obtaining slightly better scores. Additionally, a qualitative understanding of the models may be obtained by looking at the generated molecules. For instance, the image below depicts the best molecule generated by each baseline model for one of the 20 goal-directed benchmarks.
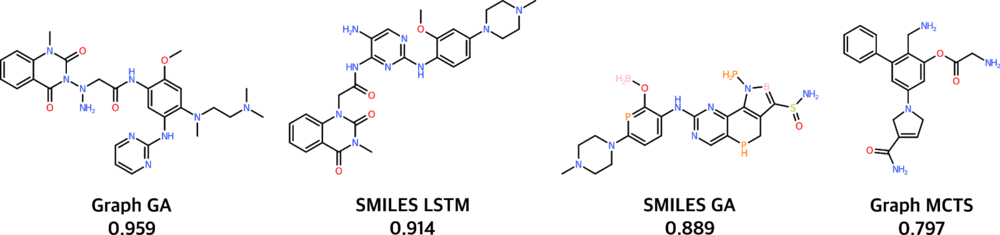
The score is given out of a maximum of 1.Best molecule generated by the considered baseline models for the Osimertinib MPO benchmark. The score is given out of a maximum of 1.The quality of the generated molecules is difficult to assess, and also difficult to incorporate in the molecule scores. The reason for this difficulty is that it is hard to encode rules of what structural features chemists consider to be unsuitable. When appraising the quality of molecules generated by the baseline models for the goal-directed tasks, nevertheless, we consider the SMILES LSTM model to be the most adequate (although its scores are slightly lower than the Graph GA model): the molecules it generates correspond better to what is commonly accepted to be drug-like.
Going forward
The assessment of models for de novo design presented in our publication is neither complete nor final: we selected only a subset of existing models for evaluation, and we could not evaluate models that have not been published yet. To keep the comparison up-to-date, and to allow others to contribute, we decided to provide a public leaderboard, which is now accessible. Researchers who develop a new model and assess it with GuacaMol are encouraged to participate in the competition by submitting their results to be displayed on this page.
The GuacaMol benchmarks are not meant to be final either. Similarly to other competitions, as models become better, the tasks tend to be too easy, and it is therefore necessary to make the benchmarks more difficult. It is therefore our intention to update the benchmark suite in the future, together with the community. It is still unclear to which extent it will be possible to incorporate the compound quality in the benchmark scores as part of this effort. We welcome feedback and suggestions from the community.
Back to blog post and videos